DNA Nanoswitch Calipers Benefit From Piezo Nanopositioning Stages
When DNA was first identified by Swiss chemist Friedrich Miescher in the 19th century, it was a mysterious biological compound, which he called “nuclein”. While Miescher may have discovered DNA originally, it was Watson and Crick who brought it to fame and captured the Nobel Prize by elucidating the double helical structure. Helped immensely by crystallographers, including the oft overlooked Rosalind Franklin, they were able to zero in on a model of DNA structure which holds today1. Who knows whether these early scientists ever considered the many applications that DNA would be useful for, all these years later.
Most of us are familiar with genetic identification for forensic and ancestry investigations, but there are numerous use cases of DNA in biochemistry and nanotechnology which receive much less attention. For example, some techniques employ DNA scaffolds to build upon or utilize DNA origami structures for biosensing and drug delivery2. These endeavors continue to show promise to enable novel research techniques as well as advance the state of medical technology, microscopy, and nanotechnology.

The P-527 nanopositioning stage on top of the U-751 long travel piezo motor stage inside the microscope. (Source: The Wyss Institute)
While nano is often synonymous with semiconductor chips, there are plenty of other areas of focus. Consider DNA with approximately a 2nm diameter and 3.4nm distance between full turns. Along with being highly programable, this natural molecule has become one of the best tools to understanding biological matter at the nano scale. PI has been instrumental in this field, supplying solutions such as piezo flexure stages which are capable of even sub nanometer precision in some cases.

The P-527 multi-axis piezo nanopositioning stage provides 200µm travel in X and Y and 20µm in Z. The maximum resolution is 0.1 nanometer in Z and 0.5 nanometer in both X and Y. The flexure guided design with direct-metrology position feedback provides backlash-free motion with very high dynamics, linearity, and position stability.
While it is it doubtful that the early researchers ever considered a future where this molecule would be used as a mechanical measurement tool, that is exactly what the Molecular Robotics Initiative at Wyss Institute at Harvard University has achieved3.
A caliper is defined by Merriam-Webster as “a measuring instrument with two legs or jaws that can be adjusted to determine thickness, diameter, or distance between surfaces.” By combining particle trapping techniques, both optical and magnetic, with single molecule techniques, the Wyss team and collaborators have created what may be the world’s smallest measurement caliper.
Video: DNA Nanoswitch Calipers (Source: The Wyss Institute)
The tool, named “DNA Nanoswitch Caliper,” or DNC, uses a hairpin DNA structure. This molecule is tethered between microparticles with sequence specific bounding. Utilizing the programmable nature of DNA allows the tether points to be designed with a set binding force. Trapping techniques can apply this force and stretch the hairpin to its fully elongated state.
Additional sections of the hairpin are programmed to bind to the biomolecule of interest, at different locations and with different forces. As such, repeated applied forces create an opening and closing of the DNC “legs” which can be correlated to distance in the molecule being measured.
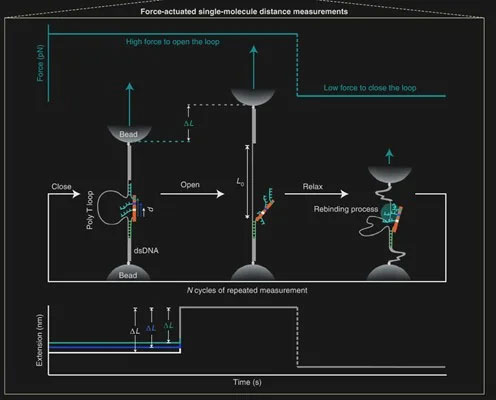
Force actuated single-molecule distance measurement principle (Source: Nature Nanotechnology)
Remarkably, the piconewton forces and nanometer distances are measured with enough statistical accuracy for even fingerprint identification of DNA and protein targets. In turn, DNCs should be an excellent tool for elucidating structural information in proteomic studies.
While DNA sequencing has become ubiquitous and straightforward, protein sequencing and structural elucidation is not as simple. The larger structural possibilities, different folding processes, and environmental interactions add to the complications. Techniques like x-ray diffraction often require crystallization outside of native environments and provide only a snapshot in time. Further analysis by single molecule techniques and simulations are often required to provide a dynamic understanding of protein structure and function. Since DNCs provide a method to make extremely precise distance measurements at this scale, they can be used to resolve previously unanswered questions in biochemistry.
It will be exciting to see where DNCs make impacts in the coming years. Rest assured - PI will be around to offer advanced technologies to improve automation and precision in these applications. At PI, we are dedicated to enabling advancements in nanotechnology and medicine by providing the highest quality of motion devices as well as advanced control electronics and amplifiers.
1.www.nature.com/scitable/topicpage/discovery-of-dna-structure-and-function-watson-397/
2.wyss.harvard.edu/technology/dna-nanotechnology-tools-from-design-to-applications/
3.wyss.harvard.edu/news/fingerprinting-proteins-with-force/
